Microarray
Advertisement
PheMaDB (Phenotype Microarray DataBase) v.1.1
PheMaDB is a web-based data management system to store and analyze OmniLog Phenotype Microarray data.
Advertisement
GNomEx v.5.3.2
GNomEx is Genomic LIMS and Analysis Project Center designed by the Huntsman Cancer Institute for use by microarray and next generation sequencing core facilities and large research laboratories.
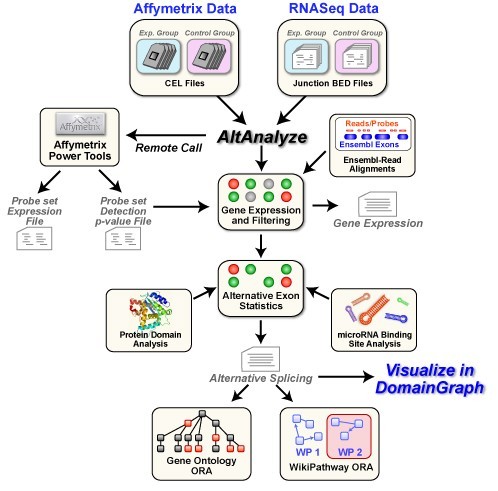
AltAnalyze for Linux 2.02 Beta v.1.0
AltAnalyze is a freely available, open-source and cross-platform program that allows you to take RNASeq or relatively raw microarray data (CEL files or normalized), identify predicted alternative splicing or alternative promoter changes and view how
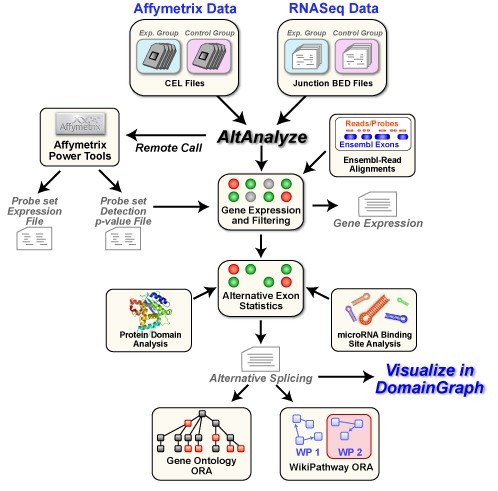
AltAnalyze for Mac OS X 2.02 Beta v.1.0
AltAnalyze is a freely available, open-source and cross-platform program that allows you to take RNASeq or relatively raw microarray data (CEL files or normalized), identify predicted alternative splicing or alternative promoter changes and view how
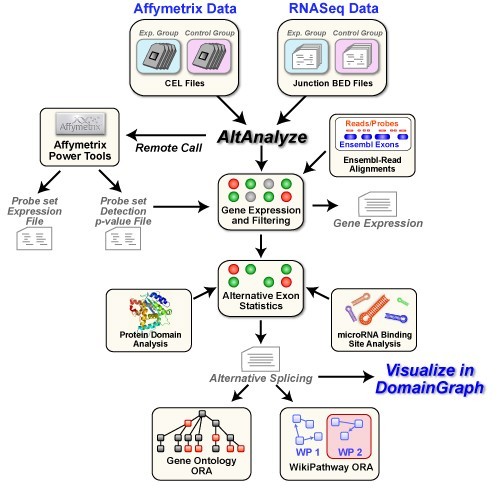
AltAnalyze x64 2.02 Beta v.1.0
AltAnalyze is a freely available, open-source and cross-platform program that allows you to take RNASeq or relatively raw microarray data (CEL files or normalized), identify predicted alternative splicing or alternative promoter changes and view how
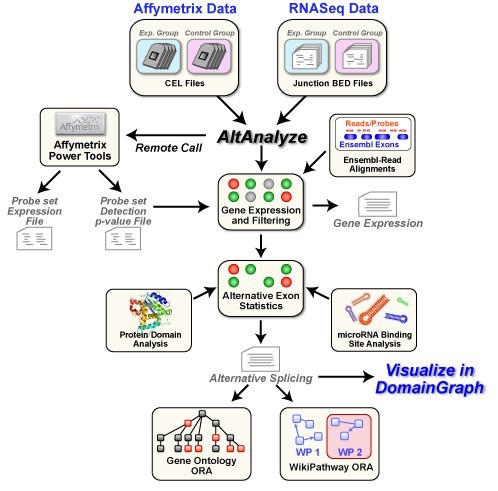
AltAnalyze 2.02 Beta v.1.0
AltAnalyze is a freely available, open-source and cross-platform program that allows you to take RNASeq or relatively raw microarray data (CEL files or normalized), identify predicted alternative splicing or alternative promoter changes and view how
MeV for Mac OS X 4.6.2 r2536 v.2536
MeV or Multiple Experiment Viewer is a Java tool to help you with microarray gene expression data analysis.
MeV 4.6.2 r2536 v.2536
MeV or Multiple Experiment Viewer is a Java tool to help you with microarray gene expression data analysis.
BMDExpress v.1 4
BMDExpress is a Java application used to analyze dose-response data from microarray experiments.
SNP HiTLink v.1. 1. 2000
During this recent decade, microarray-based single nucleotide polymorphism (SNP) data are becoming more widely used as markers for linkage analysis in the identification of loci for disease-associated genes.
PeakFinder v.1.0
Locates peaks (cohesin binding sites) in yeast ChIP microarry data. PeakFinder software was developed to find cohesin binding sites represented by the peaks in yeast chromatin immunoprecipitation (ChIP) microarray data,